Álvaro Ramírez
View DICOM files from your X-ray
Got a CD with my chest X-ray from the hospital. Was expecting a pdf or an image of sorts, but the CD content was rather different. For starters, it was targeted at Windows users (AUTORUN.INF, MediaViewerLauncher.EXE and a bunch of DLLs):
$ find . -exec file --mime-type '{}' \;
./AUTORUN.INF: text/plain
./DICOMDIR: application/dicom
./MediaViewerLauncher.EXE: application/octet-stream
...
./Libraries/BASEPRINTER.DLL: application/octet-stream
./Libraries/CDDATABURNER.DLL: application/octet-stream
./Libraries/COM.DLL: application/octet-stream
...
./Libraries/ACE.DLL: application/octet-stream
./Libraries/ACE_SSL.DLL: application/octet-stream
./Libraries/ATL90.DLL: application/octet-stream
...
./DICOM/PAT_0000: application/x-directory
./DICOM/PAT_0000/STD_0000/SER_0000/OBJ_0001/IM_0001: application/dicom
./DICOM/PAT_0000/STD_0000/SER_0001/OBJ_0001/ED_0001: application/dicom
./DICOM/PAT_0000/STD_0000/SER_0002/OBJ_0001/ED_0001: application/dicom
./Worklist/ClinicalInfo/067eccde-b299-e511-9114-005056ad3afe.mht: text/html
./Worklist/Report/067eccde-b299-e511-9114-005056ad3afe.mht: text/html
./Worklist/Worklist.wl: application/octet-stream
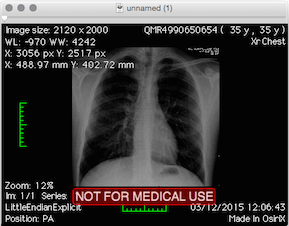
I'm on a Mac, so most of these files were not useful to me. The more interesting files were IM_0001 and ED_0001 with "application/dicom" MIME type. DICOM files stand for Digital Imaging and Communications in Medicine. How to view these on a Mac? OsiriX viewer is an option. OsiriX, though on the heavy side (100.7MB download), it rendered the X-ray successfully.
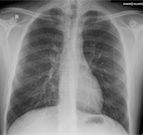
Unsurprisingly, ImageMagick's convert utility also handles DICOM files. Converting to PNG worked well.
$ convert ./DICOM/PAT_0000/STD_0000/SER_0001/OBJ_0001/ED_0001 ED_0001.png
DICOM files also hold patient's metadata and optional reports. The file format is well known. OsiriX gives you access to it, but a few lines of python can also extract it for you. First install the pydicom package:
$ sudo pip install pydicom
Running the python interpreter is enough to peak at the metadata:
>>> import dicom >>> ds = dicom.read_file("./DICOM/PAT_0000/STD_0000/SER_0000/OBJ_0001/IM_0001") >>> ds
(0008, 0000) Group Length UL: 400 (0008, 0005) Specific Character Set CS: 'ISO_IR 100' (0008, 0016) SOP Class UID UI: Computed Radiography Image Storage (0008, 0020) Study Date DA: '20151203' (0008, 0021) Series Date DA: '20151203' (0008, 0023) Content Date DA: '20151203' (0008, 0030) Study Time TM: '120519.000000' (0008, 0031) Series Time TM: '120520.000000' (0008, 0033) Content Time TM: '120643.000000' (0008, 0060) Modality CS: 'CR' (0008, 0070) Manufacturer LO: 'Canon Inc.' ...
There were other DICOM files with a report:
>>> import dicom >>> ds = dicom.read_file("./DICOM/PAT_0000/STD_0000/SER_0001/OBJ_0001/ED_0001") >>> ds
(0008, 0005) Specific Character Set CS: 'ISO_IR 100' (0008, 0016) SOP Class UID UI: Encapsulated PDF Storage ... (0042, 0012) MIME Type of Encapsulated Document LO: 'application/pdf'
DCMTK is another alternative tool to extract DICOM metadata. The source is available and can be built:
$ tar xf dcmtk-3.6.0.tar.gz $ cd dcmtk-3.6.0 $ cmake . $ make
Or installed via homebrew:
$ brew install dcmtk
DCMTK includes dcmdump. You can use it to dump DICOM files:
$ dcmdata/apps/dcmdump DICOM/PAT_0000/STD_0000/SER_0000/OBJ_0001/IM_0001
# Dicom-File-Format # Dicom-Meta-Information-Header # Used TransferSyntax: Little Endian Explicit (0002,0000) UL 192 # 4, 1 FileMetaInformationGroupLength (0002,0001) OB 01\00 # 2, 1 FileMetaInformationVersion (0002,0002) UI =ComputedRadiographyImageStorage # 26, 1 MediaStorageSOPClassUID (0002,0003) UI [1.2.392.200046.100.2.1.1.42667.20151203120519.1.1.1] # 52, 1 MediaStorageSOPInstanceUID (0002,0010) UI =LittleEndianExplicit # 20, 1 TransferSyntaxUID (0002,0012) UI [1.3.46.670589.42.1.4.4.5] # 24, 1 ImplementationClassUID (0002,0013) SH [PhilipsISPACS445] # 16, 1 ImplementationVersionName ...
Of interest, David Clunie's Medical Image Format Site.